Troubleshooting
Charles Carey
09 May, 2018
Source:vignettes/troubleshooting.Rmd
troubleshooting.Rmd
Troubleshooting.
If we encounter problems, such as an empty result, we can always ask for Monarch’s response to our query.
Response from Monarch Initiative.
library(monarchr)
gene <- "NCBIGene:8314"
results <- bioentity_homologs(gene)We returned the response as a monarch_api (S3) class with its own print method
class(results$response)## [1] "monarch_api"The print method only shows the URL and highest 2 levels of the contents.
results$response## <monarch https://api.monarchinitiative.org/api/bioentity/gene/NCBIGene%3A8314/homologs/?rows=100&fetch_objects=true&format=json>
## <Showing parsed R objects from json response. 2 level(s) deep>
## List of 7
## $ compact_associations: NULL
## $ associations :'data.frame': 17 obs. of 14 variables:
## ..$ provided_by :List of 17
## ..$ relation :'data.frame': 17 obs. of 2 variables:
## ..$ object_extensions : logi [1:17] NA NA NA NA NA NA ...
## ..$ publications :List of 17
## ..$ object :'data.frame': 17 obs. of 3 variables:
## ..$ negated : logi [1:17] FALSE FALSE FALSE FALSE FALSE FALSE ...
## ..$ slim : logi [1:17] NA NA NA NA NA NA ...
## ..$ subject_extensions: logi [1:17] NA NA NA NA NA NA ...
## ..$ type : logi [1:17] NA NA NA NA NA NA ...
## ..$ qualifiers : logi [1:17] NA NA NA NA NA NA ...
## ..$ evidence_graph :'data.frame': 17 obs. of 2 variables:
## ..$ evidence_types : logi [1:17] NA NA NA NA NA NA ...
## ..$ id : chr [1:17] "beb2860f-b6ab-472e-8c77-eb8ff2a5da99" "47931d07-86d6-40a2-82d8-d7919a0e8ad5" "5eea0259-b923-49f5-a344-86820defa027" "bfe3507a-f8bf-4a7d-949b-ab3e63dd8e95" ...
## ..$ subject :'data.frame': 17 obs. of 3 variables:
## $ objects : chr [1:17] "FlyBase:FBgn0262166" "HGNC:12513" "HGNC:12515" "HGNC:19678" ...
## $ highlighting : NULL
## $ numFound : NULL
## $ facet_counts :List of 2
## ..$ subject_taxon_label:List of 1
## ..$ object_closure :List of 25
## $ docs : NULLTo view at an alternative depth, supply the depth setting to the print method.
# print(results$response, depth=3) # not run.Troubleshooting the server status
class(results$response$response) # this is the response we saved from the call to httr GET(url)## [1] "response"results$response$url## [1] "https://api.monarchinitiative.org/api/bioentity/gene/NCBIGene%3A8314/homologs/?rows=100&fetch_objects=true&format=json"results$response$response$status_code## [1] 200#str(results$response$response)
#results$response$response$headers
#results$response$response$requestThe server should have responded with an OK (200) status. If not, we can confirm the URL ourselves by pasting into a browser, or look at the response and response headers for more info (courtesy of httr).
Viewing JSON directly from the server.
We can use the URL at the top of monarchr’s summary of the response to see the JSON results directly from the server.
(this is easier to view in Firefox):
Viewing HTML on server.
If you prefer to see the expected results as HTML, note how the example of looking up NCBIGene:8314 on Monarch Initiative website ended up at a table for Monarch’s preferred ID (HGNC:950).
[https://monarchinitiative.org/gene/HGNC:950#homologs]
Replacing HGNC:950 with NCBIGene ID, we are successfully directed to the same view.
Verifying your search returns something consistent with searching Monarch Initiative’s website directly.
- Go to Monarch’s front page: [https://monarchinitiative.org/]
- In the search box, enter and search for: NCBIGene:8314.
- Select the exact match (of thousands) from Homo sapiens. (This is the first match and shows the gene name is BAP1.)
- Select homologs from the tabs at the top of the next page.
You should see something similar to the following view.
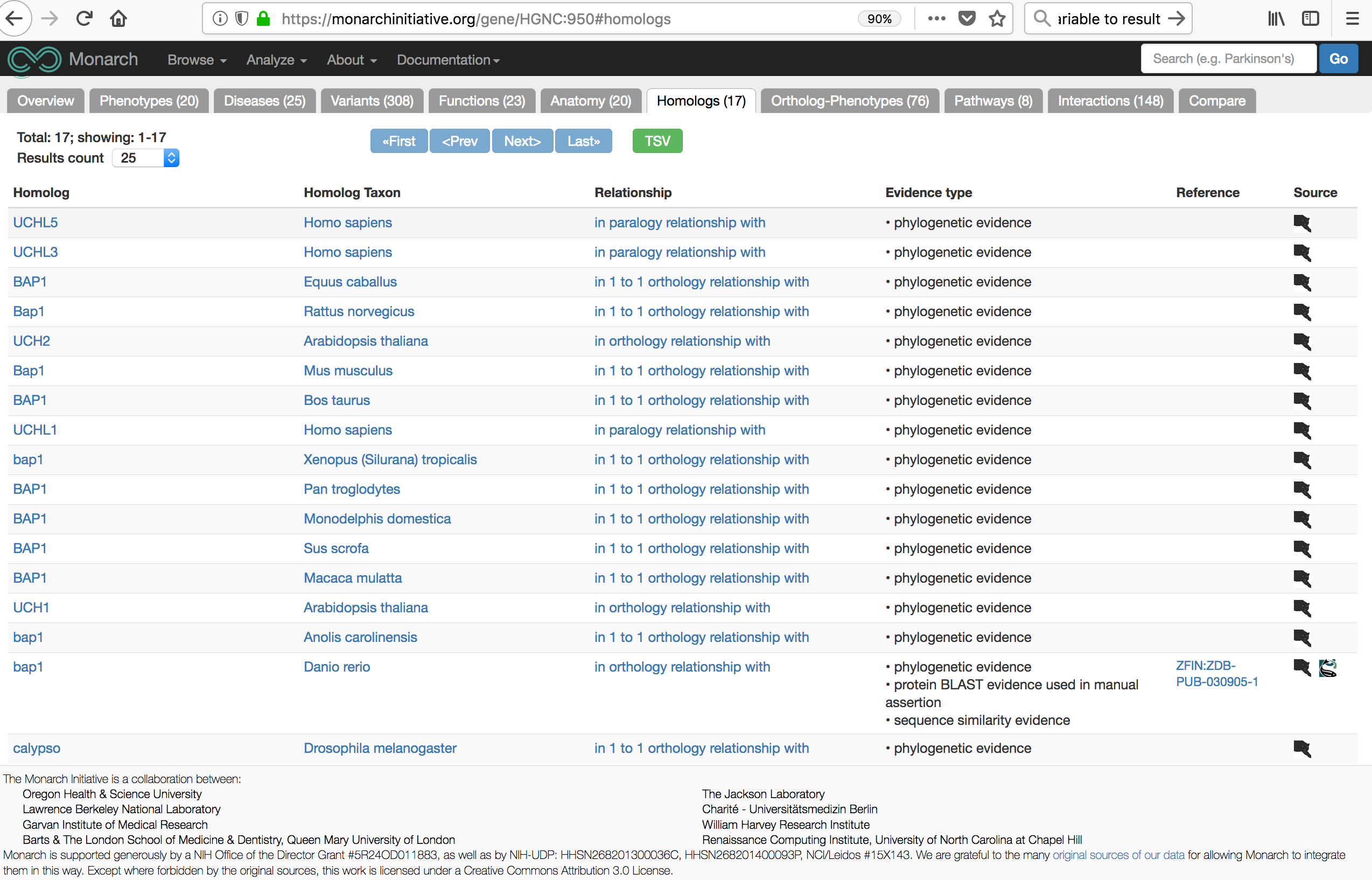
Homologs as seen on Monarch Inititiative Website.
In this case, the results shoud be consistent with what we see below.
results$homologs## # A tibble: 17 x 10
## subject.taxon subject.id subject relation object object.id object.taxon
## * <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 Homo sapiens HGNC:950 BAP1 in para… UCHL5 HGNC:196… Homo sapiens
## 2 Homo sapiens HGNC:950 BAP1 in para… UCHL3 HGNC:125… Homo sapiens
## 3 Homo sapiens HGNC:950 BAP1 in 1 to… BAP1 NCBIGene… Equus cabal…
## 4 Homo sapiens HGNC:950 BAP1 in 1 to… Bap1 RGD:1311… Rattus norv…
## 5 Homo sapiens HGNC:950 BAP1 in orth… UCH2 NCBIGene… Arabidopsis…
## 6 Homo sapiens HGNC:950 BAP1 in 1 to… Bap1 MGI:1206… Mus musculus
## 7 Homo sapiens HGNC:950 BAP1 in 1 to… BAP1 NCBIGene… Bos taurus
## 8 Homo sapiens HGNC:950 BAP1 in para… UCHL1 HGNC:125… Homo sapiens
## 9 Homo sapiens HGNC:950 BAP1 in 1 to… bap1 Xenbase:… Xenopus (Si…
## 10 Homo sapiens HGNC:950 BAP1 in 1 to… BAP1 NCBIGene… Pan troglod…
## 11 Homo sapiens HGNC:950 BAP1 in 1 to… BAP1 NCBIGene… Monodelphis…
## 12 Homo sapiens HGNC:950 BAP1 in 1 to… BAP1 NCBIGene… Sus scrofa
## 13 Homo sapiens HGNC:950 BAP1 in 1 to… BAP1 NCBIGene… Macaca mula…
## 14 Homo sapiens HGNC:950 BAP1 in orth… UCH1 NCBIGene… Arabidopsis…
## 15 Homo sapiens HGNC:950 BAP1 in 1 to… bap1 NCBIGene… Anolis caro…
## 16 Homo sapiens HGNC:950 BAP1 in orth… bap1 ZFIN:ZDB… Danio rerio
## 17 Homo sapiens HGNC:950 BAP1 in 1 to… calyp… FlyBase:… Drosophila …
## # ... with 3 more variables: evidence <fct>, publications <fct>,
## # provided_by <fct>